Genomics research and analyses prompt researchers and scientists to seek highly optimized computing and storage solutions to handle an immense volume of next generation sequencing (NGS) sequence data. Oracle Cloud Infrastructure (OCI) provides accelerated solutions to meet the challenges with cost-effectiveness, high performance, and availability. Genome Analysis Toolkit (GATK) is an analysis toolkit commonly used by most genomic research and clinical analyses.
The OCI solution team recently created an optimal solution for the GATK best practice pipeline that reduced job runtime to record-breaking 2.23 hours on a two-socket Intel system, which represented a near 10-times speed up from the run time using baseline run script that came with the GATK code. The OCI solution provides automated optimization on secondary analysis sequence alignment, marking duplicates, variant discovery, and variant filtering on all OCI Intel-based systems. The solution is highly adjustable to meet specific user requirements and can be easily integrated into customer’s current solutions. It’s readily available to OCI customers on request.
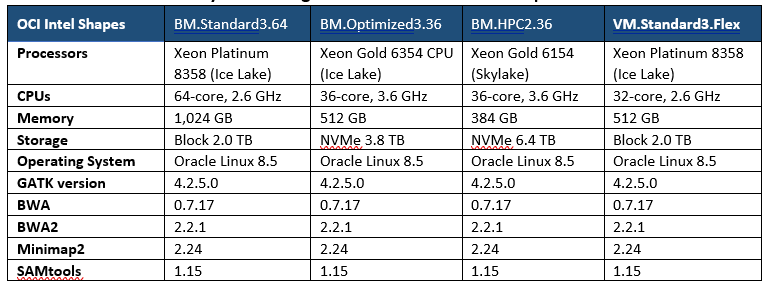
OCI Intel shapes
OCI provides a high-performance environment with cost-effective computing and optimal performance for genomic analytic solutions. OCI solution experts assist customers to customize and tune their workloads to obtain the best performance on the OCI systems. In this blog, we demonstrate the optimized solution using GATK’s best practices workflow for Germline short variant discovery to accelerate the run time by nearly 10 times, while retaining the high accuracy of the outputs.
OCI offers various Intel-based shapes (platforms) to meet customer needs. We deployed three bare metal and one standard virtual machine (VM) instances for performance benchmarking. The configurations of each instance are listed in Table 1:
Table 1. OCI benchmark system configurations and GATK version specification
The OCI genomics solution applies the power of high-performance computing and data storage for NGS data analysis on cloud by providing the following capabilities:
- High performance and scalability: OCI brings powerful, high-performance computing (HPC) capabilities to solve complex mathematical and scientific problems across industries. OCI’s bare metal servers, BM.Optimized3.36 and BM.HPC2.36, are connected with Oracle’s cluster networking, providing access to ultra-low latency RDMA network.
- Economics, flexibility, and availability: OCI brings consumption-based costs, flexible computing to meet customers’ budgets, while offering on-demand potential to scale tens of thousands of cores simultaneously. OCI provides tools to automate and run jobs seamlessly.
- Security and governance: OCI is a security-first public cloud infrastructure that Oracle built for enterprise critical workloads. Essential security services complement the workloads to provide the required levels of security for your most business-critical workloads.
Genome analysis pipelines
The Genome Analysis Toolkit (GATK), developed by Broad Institute, is widely used by biologists free of charge. GATK consists of a set of tools for Germline and cancer genomic analyses. The best practice pipelines, published by Broad Institute, are often adopted by the genomics community. GATK code is bundled with a run script (baseline script), which runs most GATK tools using either a single thread or multithreads.
However, the baseline script often delivers poor scalability if it’s not tuned to match the compute node. Our baseline benchmark test ran for more than 20 hours to complete a pipeline analysis from alignment to produce gene variants calling for a 30x coverage whole genomic sequences (WGS) data set (See Figure 2). Fortunately, you can create solutions by splitting the workload into sequence intervals for speeding up runtime concurrently with GATK4 tools.
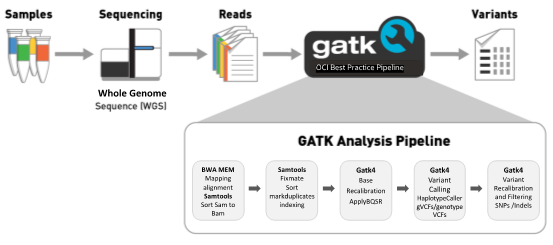
This blog uses the GATK best practices workflow for Germline short variant discovery as a case study to optimize the GATK4 workload on OCI’s Intel shapes. The whole genome sequencing data set NA12878 with 30x coverage was downloaded using Google Cloud Utility. The dataset, HG002.hiseqx.pcr-free.30x.R[1-2].fastq.gz, contains 777,447,412 reads in paired-end with 150 bps in length per read. The reference genome and known variant call format (VCF) reference files are obtained from the Broad Institute resource bundle. The reference genome Grch38/Hg38, including 1,000 genomes’ known VCFs, was downloaded from the Broad Institute FTP server. The scripts used for step-by-step processes are described here.
OCI Optimized GATK best practice pipeline
As described, the original baseline run script provided with GATK code doesn’t perform well, but it can be improved by tuning the source code, enabling multithreads, splitting input intervals, and selecting newer, faster tools in the pipeline without losing output accuracy. OCI’s optimizations include the following features applied to the original baseline run script:
- Optimizing sequence alignment: BWA is a tool uses in the first step of the pipeline for sequence mapping. While we can improve its performance with concurrent memory allocation and simultaneously multithreading, the faster alternative tools, such BWA mem2 and minimap2, which are implemented with AVX51 extension, can replace BWA to speed up the alignment process. Process binding can also improve the performance.
- Improving sorting and marking duplicates with SAMtools: Picard marking duplicates are a single-threaded process, where SAMtools with the optional ‘-s’ mode produces identical output to the Picard tool.
- Running GATK with splitting intervals: Programs in the pipeline, such as BaseRecalibration, ApplyVQSR, Haplotypecaller, and GenotypeVCF, can run with splitting intervals to run multiple intervals concurrently. The outputs of each interval are then combined.
- Variant calling with PairHMM and Smith-Waterman: A variant calling tool using Haplotypecaller is implemented with faster algorithms, such as
PairHMM and Smith-Waterman. Setting options with --pairhmm and --smith-waterman to FASTEST_AVAILABLE and --native-pair-hmm-threads 4significantly speed up the performance.
Tuning Java options: Java’s garbage collection tuning improves the runtime performance. These options include -Xmx4G, -XX:+UseParallelGC, and -XX:ParallelGCThreads=#threads.
GATK4 benchmark performance on OCI Intel shapes
Speed up 10 times from standard baseline run
The results of the tuning and optimization described in Section C are shown in Figure 2. Our optimized script is combined with alignment with minimap2, marking duplicates with SAMtools, splitting intervals and running concurrently in programs of Baserecalibration, ApplyBQSR, Haplotypecaller, and GenotypeVCF. The pipeline running the 30x coverage WGS dataset using the optimized script completed in 2.23 hours on a 64-core Ice Lake system (Standard3.64), which is almost 10 times faster than the baseline script. The resulting VCF file is 97.5% in concordance with the original pipeline script.

Performance comparison of GATK sequence alignment tools
Typically, the GATK best practice pipeline spends more than 80% of its runtime in sequence alignment. Three generations of sequence alignment tools—BWA, BWA-MEM2, and Minimap2—can be found from GATK, and they all generate nearly identical alignment outputs. This exercise evaluated the performance of these three tools using OCI’s optimized script on OCI’s bare metal and VM shapes. BWAMEM2 demonstrated noticeable performance improvement because it was accelerated with AVX512. (I added this sentence as a response to Marissa’s suggestion.) The results also showed that the 64-core standard bare metal shape with Ice Lake has the best performance among all shapes (Figure 3). The chart also showed a 17% performance improvement between latest two generations of Intel processors (Ice Lake in BM.Optimized.36 versus Sky Lake in BM.HPC2.36).

Performance of the entire GATK best practice pipeline
The GATK Germline best practice pipeline starts with sequence alignment or mapping to the reference genome and ends with variant (SNPs and indels) recalibration and filtering. The total runtime of OCI optimized scripts ranges 2.86–5.74 hours on the four Intel shapes. These results are by far the best performance achieved by using GATK tools on OCI Intel Xeon-based systems, as shown in Figure 4.

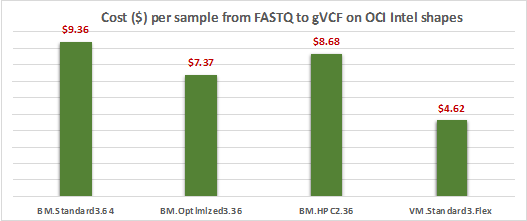
Cost of running a 30x coverage WGS sample on OCI Intel shapes
The GATK Germline best practice pipeline uses no more than 128 GB of memory and 1 TB of storage to analyze 30x coverage WGS dataset. The cost of running the pipeline per sample is calculated by multiplying the cost per hour of machine with runtime of the job. The chart in Figure 5 shows that VM shapes, which were configured with minimum required memory, is cheapest to get the job done. Bare metal shapes cost more because they were configured with more memory and other hardware unused by GATK and users didn’t adjust their configurations. In comparison, the machine with the most processors, namely BM.Standard3.64 was the most expensive, but it finished the job fastest.
Closing remarks
The Oracle Cloud Infrastructure optimized run script for the GATK best practice pipeline achieved 10 times the throughput performance speed, compared to the baseline run script that came with GATK code. The system with most processor cores (BM.Standard3.64) runs the fastest among all four evaluated shapes, but the cost of the job run was the highest. In comparison, the VM system with flex configuration, VM.Standard3.Flex, get the job done at the lowest cost. Customers can use this guideline to help them select the optimal shape for their sequencing workload.
For customers who routinely process large batches of data samples, they can also use the benchmark data in Figure 4 to estimate workload throughput performance. For a 32-node cluster, it takes less than three days to complete analysis of 1,000 samples with 30x coverage whole human genome reads.
References
2 “Introduction to the GATK Best Practices.” Broad Institute, January 9, 2018. https://software.broadinstitute.org/gatk/best-practices
5 Jacob R. Heldenbrand, Saurabh Baheti, Matthew A. Bockol et al. “Performance benchmarking of GATK3.8 and GATK4.” Mayo Clinic and Illinois Strategic Alliance for Technology-Based Healthcare, June 2018. https://www.biorxiv.org/content/biorxiv/early/2018/06/18/348565.full.pdf


