Humanity has witnessed an unprecedented crisis over the past months. With the emergence of the COVID-19 pandemic, Life Sciences researchers across the globe have been working tirelessly to find a treatment for the disease. Oracle Cloud Infrastructure high-performance computing (HPC) is playing its part by enabling researchers with access to cutting-edge hardware.
Scalable HPC infrastructure on Oracle Cloud Infrastructure (OCI) significantly speeds up the drug discovery process. OCI has supported researchers across the globe in their efforts to find a treatment for COVID-19, including the Brazilian Biosciences National Laboratory (LNBio). Researchers at LNBio have made advancements in the fight against COVID-19 through Oracle for Research, a new program that provides Oracle Cloud credits and related resources to projects that drive meaningful change in the world.
A Race for a Cure
Given the prevailing situations, researchers are in a race to find a COVID-19 vaccine and treatment. The drug discovery life cycle from the beginning of the project to regulatory approvals takes over 12 years. Also, only one out of 5,000 experimental drug molecules are approved for safe human usage. With COVID-19, time is a luxury that the researchers don’t have.
Keeping this fact in mind, the researchers at LNBio are attempting to follow a drug-repurposing approach. In this approach, a medicine pre-approved by the regulatory agencies for treatment of another disease is tested for treatment of a newly emerging disease. This approach may significantly cut down the time it takes to make a treatment available to the larger population.
The team of researchers at LNBio and the Brazilian Center for Research in Energy and Materials (CNPEM), part of the Brazilian Ministry of Science, Technology and Innovation, are working on identifying drug molecules, which attach to viral proteins and stop the virus from causing infection or multiplying inside the human body (Figure 1). This process can help patients to recover from COVID-19, because it helps the immune system to fight the infection by decreasing the number of viral particles inside the patient’s body.
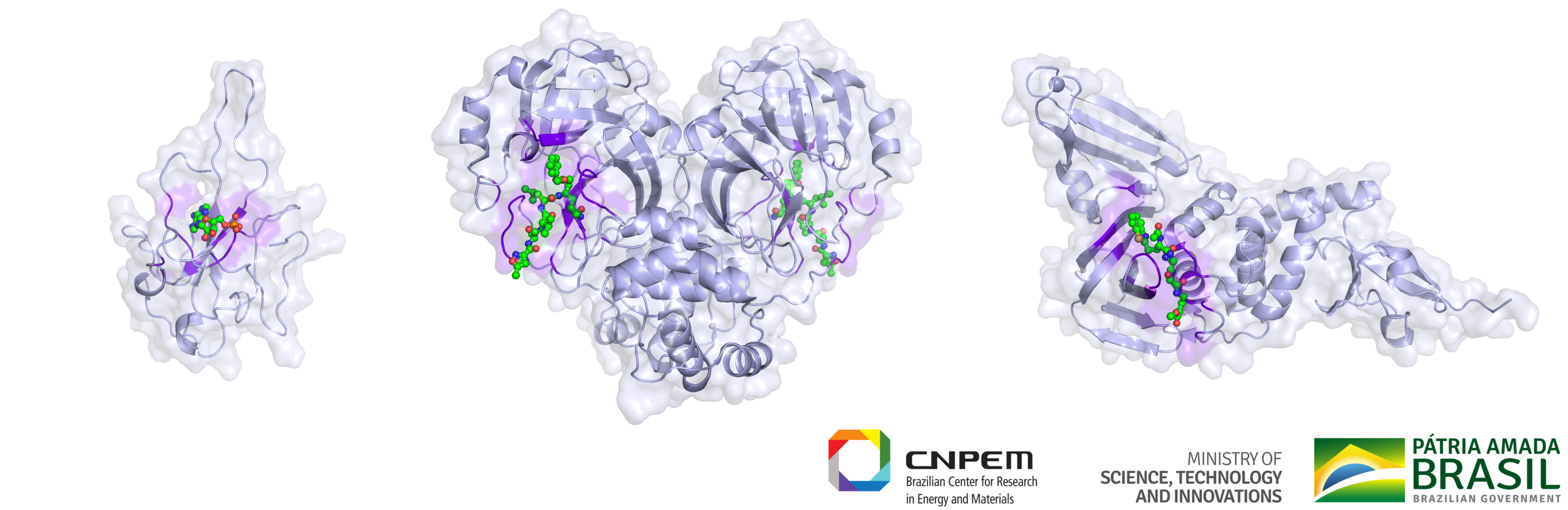
Figure 1. Researchers at the Brazilian Biosciences National Laboratory (LNBio) and the Brazilian Center for Research in Energy and Materials (CNPEM) are targeting active sites (from left to right) nucleocapsid, 3CL-protease, and PL-protease. Drug molecules blocking these active sites stop the virus from causing infection or multiplying inside our body. (Courtesy: LNBio)
Process of Drug Repurposing
Researchers of LNBio started with 2,097 pre-approved drug molecules available in the DrugBank database. They performed the process of virtual screening, where 419,400 conformations of these 2,097 molecules attached to the target locations on virus were generated through molecular docking algorithm. Briefly, this algorithm takes the atom Cartesian coordinates of molecules and applies random rotation in all vectors formed by rotatable bonds, generating thousands of possible conformations for the original molecule. Each conformation is rotated and translated to generate a “molecular pose” in the active site of viral protein. An empirical score function is calculated for each pose.
This process is repeated millions of times for each target. A customized ranking algorithm narrows down the prospects to 200 best poses. Finally, using these 200 poses generated through virtual screening, molecular simulations help identify three drug molecules, which have a potential of stopping multiplication of the virus (Figure 2). Solving this problem is computationally intensive and requires scalable robust infrastructure, which is often difficult for research organizations to access quickly and cost effectively.
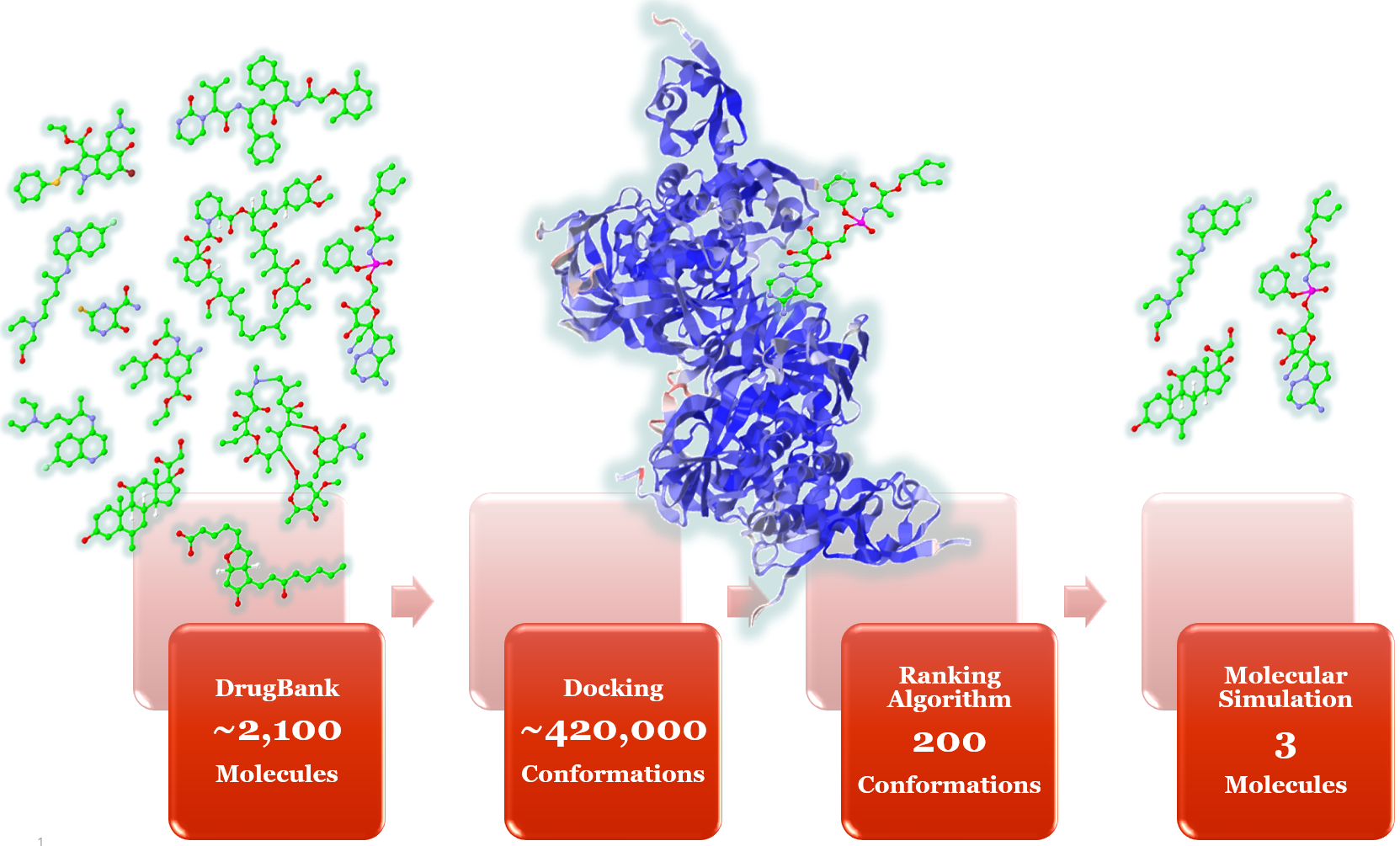
Figure 2. Drug repositioning process. Molecular structures are used for schematic purpose only, and do not reflect the actual research outcomes.
Security, Performance, and Economics
Oracle Cloud Infrastructure is built considering security and performance requirements of large enterprises. OCI bare metal HPC compute instances, networked over ultra-low-latency remote direct memory access (RDMA) with approximately 1.5-µs latency cluster networking provide an ideal solution for CPU-based, large-scale molecular simulations. Benchmarking studies have shown that OCI HPC provides the best performance in public cloud for CPU-based molecular simulations. At the same time, GPU-based algorithms also run efficiently on the Oracle Cloud Infrastructure NVIDIA Tesla V100 GPU shapes with NVLink interconnect, providing fast communication between the GPUs.
A study recently published in the Journal of the American Medical Association (JAMA) found that biopharmaceutical companies spent an estimated $985 million on new drug development and research between 2009-2018. Through OCI HPC, pharmaceutical research and development firms can help reduce their time-to-market for new drugs through improved performance, which can potentially offer huge cost savings.
Accelerated Discovery with the Scalability on Oracle Cloud Infrastructure
By taking advantage of scalability offered by GPU and CPU resources on the Oracle Cloud Infrastructure HPC, the researchers are looking at reducing the time for virtual screening by one-sixth. “We were able to reduce the time to complete our virtual screening workflow by half in some early tests. Now, by using the scalability and flexibility offered by Oracle Cloud Infrastructure, we are looking at reduction in time for virtual screening by six folds,” said Leandro Oliveira Bortot, post-doctoral researcher at LNBio and CNPEM. “We’re looking forward to availability of NVIDIA A100 GPUs on the Oracle Cloud Infrastructure, as it’s the easiest way for us to have access to the latest hardware on-demand,” he added.
Oracle Cloud Infrastructure continues to work with LNBio research team and other pharmaceutical research and development companies and to provide them with technology that can help them accelerate their drug discovery process.