There are so many interesting and potentially useful data sets available today via REST: social, financial, science, health, weather – the list goes on and on. Autonomous Database makes it really easy to integrate these sources using SQL queries – eliminating the need for intermediate processing and storage and making that data available to any SQL client.
Simplify your data architecture
Autonomous Database has rich support for using Oracle SQL to query data that is stored outside of the database. This data may be available in other databases, object stores or thru REST endpoints. The implication is that ADB’s SQL access capability can dramatically simplify your data architecture. Instead of deploying a separate application to interrogate, process and persist third party cloud service data – Autonomous Database can process that data on its own using PLSQL and SQL queries.
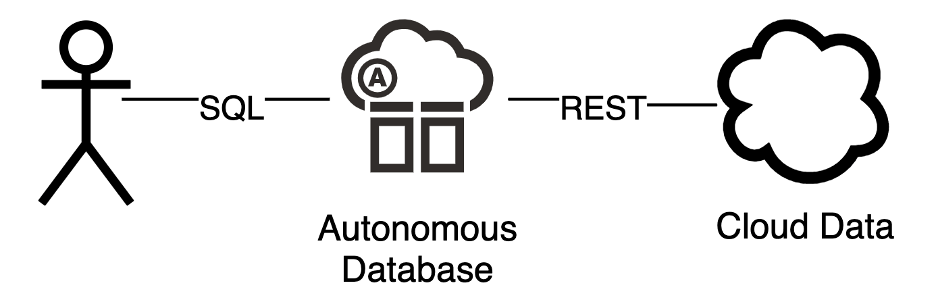
Let’s walk thru an example using the Nobel Prize REST service.
Use SQL to Query REST endpoints
We’ll go thru the following steps:
- Allow the ADB ADMIN user to query any public REST endpoint
- Create a function that queries the Nobel Prize REST endpoint
- Use that function to run a SQL query against that Nobel Prize REST endpoint
- Add structure to that query
1. Allow the ADB ADMIN user to query any public REST endpoint
One of the key benefits of Autonomous Database is that it is secure by default. As a result, the ADMIN user will update the database’s access control list (this is in addition to the OCI network’s access control list) to enable remote access via http(s). You can read more about the details in the documentation.
begin
dbms_network_acl_admin.append_host_ace(
host => '*',
ace => xs$ace_type(privilege_list => xs$name_list('http'),
principal_name => 'ADMIN',
principal_type => xs_acl.ptype_db)
);
end;
/
Here, we’re specifying that the ADMIN user can make REST calls in ADB to any external host.
2. Create a function that queries the Nobel Prize REST endpoint
Now that ADMIN can run REST calls against public endpoints, let’s write a small function that finds the Nobel Prize winners for different categories (medicine, peace, literature, etc.) by year. The REST API call that returns Nobel Prize winners for medicine from 2019 to 2021 is:
https://api.nobelprize.org/2.1/nobelPrizes?limit=1000&nobelPrizeYear=2019&yearTo=2021&nobelPrizeCategory=med
We’ll create a PLSQL function that takes arguments which mirror the Nobel Prize API:
- nobelPrizeYear – a specific year
- yearTo – used with Nobel Prize Year to specify a time range.
- nobelPrizeCategory – allows you to specify the area of interest
create or replace function nobel_prizes (
nobelPrizeYear in number default 2015,
yearTo in number default 2021,
nobelPrizeCategory in varchar2 default null
-- valid values: che, eco, lit, med, pea, phy
)
return clob is
result_row clob;
-- REST management
resp dbms_cloud_types.resp;
limit number := 1000;
params varchar2(1000);
endpoint varchar2(100) := 'https://api.nobelprize.org/2.1/nobelPrizes';
reqheader varchar2(1000) := null;
begin
-- Fill in the parameters that are unique to this endpoint
params := '?limit=' || limit
|| '&nobelPrizeYear=' || nobelPrizeYear
|| '&yearTo=' || yearTo
|| '&nobelPrizeCategory=' || nobelPrizeCategory;
-- send the request and process the result
resp := dbms_cloud.send_request(
credential_name => null,
-- headers => reqheader,
uri => endpoint || params,
method => DBMS_CLOUD.METHOD_GET,
cache => true
);
-- Get the response
result_row := dbms_cloud.get_response_text(resp);
-- Return the result
return result_row;
end;
/
There’s really only a few things about the function to call out:
- endpoint: this is the URI that targets the Nobel Prize service
- params: each REST endpoint takes a set of parameters. Here, the parameters are nobelPrizeYear, yearTo and NobelPrizeCategory
- dbms_cloud.send_request: This ADB PLSQL function does the heavy lifting. It sends and processes the request. There are some very cool things that you can do with this – including caching and viewing the details of the result
3. Run a SQL query against that Nobel Prize REST endpoint
Now that the function has been defined, let’s run a simple query. Although the query is simple – the result is a bit complicated! Don’t worry, we’ll remove that complication later.
select json_query(nobel_prizes(2019, 2021, 'med'), '$' returning clob pretty) from dual;
The SQL is using the json_query function in order to prettify the result. Here’s a portion of the printed output. Congratulations to David Julius and Ardem Patapoutian for their discoveries of receptors for temperature and touch!
{
"nobelPrizes" :
[
{
"awardYear" : "2019",
"category" :
{
"en" : "Physiology or Medicine"
},
"categoryFullName" :
{
"en" : "The Nobel Prize in Physiology or Medicine"
},
"dateAwarded" : "2019-10-07",
"prizeAmount" : 9000000,
"prizeAmountAdjusted" : 9000000,
"laureates" :
[
{
"id" : "970",
"knownName" :
{
"en" : "William G. Kaelin Jr"
},
"fullName" :
{
"en" : "William G. Kaelin Jr"
},
"motivation" :
{
"en" : "for their discoveries of how cells sense and adapt to oxygen availability",
},
{
"id" : "971",
"knownName" :
{
"en" : "Sir Peter J. Ratcliffe"
},
"fullName" :
{
"en" : "Sir Peter J. Ratcliffe"
},
"motivation" :
{
"en" : "for their discoveries of how cells sense and adapt to oxygen availability"
},
{
"id" : "972",
"knownName" :
{
"en" : "Gregg L. Semenza"
},
"fullName" :
{
"en" : "Gregg L. Semenza"
},
"motivation" :
{
"en" : "for their discoveries of how cells sense and adapt to oxygen availability"
},
}
]
},
{
"awardYear" : "2020",
"category" :
{
"en" : "Physiology or Medicine"
},
"categoryFullName" :
{
"en" : "The Nobel Prize in Physiology or Medicine"
},
"dateAwarded" : "2020-10-05",
"prizeAmount" : 10000000,
"prizeAmountAdjusted" : 10000000,
"laureates" :
[
{
"id" : "985",
"knownName" :
{
"en" : "Harvey J. Alter"
},
"fullName" :
{
"en" : "Harvey J. Alter"
},
"motivation" :
{
"en" : "for the discovery of Hepatitis C virus"
},
},
{
"id" : "986",
"knownName" :
{
"en" : "Michael Houghton"
},
"fullName" :
{
"en" : "Michael Houghton"
},
"motivation" :
{
"en" : "for the discovery of Hepatitis C virus"
},
},
{
"id" : "987",
"knownName" :
{
"en" : "Charles M. Rice"
},
"fullName" :
{
"en" : "Charles M. Rice"
},
"motivation" :
{
"en" : "for the discovery of Hepatitis C virus"
},
}
]
},
{
"awardYear" : "2021",
"category" :
{
"en" : "Physiology or Medicine"
},
"categoryFullName" :
{
"en" : "The Nobel Prize in Physiology or Medicine"
},
"dateAwarded" : "2021-10-04",
"prizeAmount" : 10000000,
"prizeAmountAdjusted" : 10000000,
"laureates" :
[
{
"id" : "997",
"knownName" :
{
"en" : "David Julius"
},
"fullName" :
{
"en" : "David Julius"
},
"motivation" :
{
"en" : "for their discoveries of receptors for temperature and touch"
},
},
{
"id" : "998",
"knownName" :
{
"en" : "Ardem Patapoutian"
},
"fullName" :
{
"en" : "Ardem Patapoutian"
},
"motivation" :
{
"en" : "for their discoveries of receptors for temperature and touch"
},
}
]
}
],
"meta" :
{
"offset" : 0,
"limit" : 1000,
"nobelPrizeYear" : 2019,
"yearTo" : 2021,
"nobelPrizeCategory" : "med",
"count" : 3,
"terms" : "https://www.nobelprize.org/about/terms-of-use-for-api-nobelprize-org-and-data-nobelprize-org/",
"license" : "https://www.nobelprize.org/about/terms-of-use-for-api-nobelprize-org-and-data-nobelprize-org/#licence",
"disclaimer" : "https://www.nobelprize.org/about/terms-of-use-for-api-nobelprize-org-and-data-nobelprize-org/#disclaimer"
},
"links" :
{
"first" : "https://masterdataapi.nobelprize.org/2.1/nobelPrizes?offset=0&limit=1000&nobelPrizeYear=2019&yearTo=2021&nobelPrizeCategory=med",
"self" : "https://masterdataapi.nobelprize.org/2.1/nobelPrizes?offset=0&limit=1000&nobelPrizeYear=2019&yearTo=2021&nobelPrizeCategory=med",
"last" : "https://masterdataapi.nobelprize.org/2.1/nobelPrizes?offset=0&limit=1000&nobelPrizeYear=2019&yearTo=2021&nobelPrizeCategory=med"
}
}
4. Add structure to that query
I removed a lot of tags in an attempt to make the output cleaner. JSON is really useful because it can be interpreted by so many different tools and languages. But, it’s not necessarily concise ;).
This result is a single JSON record – an array of the Nobel Prizes by year. Within each year is yet another array: the Laureates that won the awards. Let’s tackle the first array. We’ll turn each array value into a row – and transform a subset of the array’s attributes into columns. JSON_TABLE is the SQL function that will be used to accomplish the task:
with prizes as (
-- make the REST call
select nobel_prizes(2015, 2021, 'med') as data
from dual
)
-- Select from the “with” clause – i.e. the REST call
select award_year,
date_awarded,
prize_amount,
category,
laureates
from prizes p,
json_table(p.data, '$.nobelPrizes[*]'
columns (
award_year varchar2(4) path '$.awardYear',
date_awarded varchar2(12) path '$.dateAwarded',
prize_amount number path '$.prizeAmount',
category varchar2(100) path '$.categoryFullName.en',
laureates clob format json path '$.laureates'
)
) jt
;
| AWARD_YEAR | DATE_AWARDED | PRIZE_AMOUNT | CATEGORY | LAUREATES |
|---|---|---|---|---|
| 2019 | 2019-10-07 | 9000000 | The Nobel Prize in Physiology or Medicine | [{“id”:”970″,”knownName”:{“en”:”William G. Kaelin Jr”},”fullName”:{“en”:”William G. Kaelin Jr”},”portion”:”1/3″,”sortOrder”:”1″,”motivation”:{“en”:”for their discoveries of how cells sense and adapt to oxygen availability”,”se”:”för deras upptäckter av hur celler känner av och anpassar sig efter syretillgång”},”links”:[{“rel”:”laureate”,”href”:”https://api.nobelprize.org/2/laureate/970″,”action”:”GET”,”types”:”application/json”}]},{“id”:”971″,”knownName”:{“en”:”Sir Peter J. Ratcliffe”},”fullName”:{“en”:”Sir Peter J. Ratcliffe”},”portion”:”1/3″,”sortOrder”:”2″,”motivation”:{“en”:”for their discoveries of how cells sense and adapt to oxygen availability”,”se”:”för deras upptäckter av hur celler känner av och anpassar sig efter syretillgång”},”links”:[{“rel”:”laureate”,”href”:”https://api.nobelprize.org/2/laureate/971″,”action”:”GET”,”types”:”application/json”}]},{“id”:”972″,”knownName”:{“en”:”Gregg L. Semenza”},”fullName”:{“en”:”Gregg L. Semenza”},”portion”:”1/3″,”sortOrder”:”3″,”motivation”:{“en”:”for their discoveries of how cells sense and adapt to oxygen availability”,”se”:”för deras upptäckter av hur celler känner av och anpassar sig efter syretillgång”},”links”:[{“rel”:”laureate”,”href”:”https://api.nobelprize.org/2/laureate/972″,”action”:”GET”,”types”:”application/json”}]}] |
| 2020 | 2020-10-05 | 10000000 | The Nobel Prize in Physiology or Medicine | [{“id”:”985″,”knownName”:{“en”:”Harvey J. Alter”},”fullName”:{“en”:”Harvey J. Alter”},”portion”:”1/3″,”sortOrder”:”1″,”motivation”:{“en”:”for the discovery of Hepatitis C virus”,”se”:”för upptäckten av hepatit C-virus”},”links”:[{“rel”:”laureate”,”href”:”https://api.nobelprize.org/2/laureate/985″,”action”:”GET”,”types”:”application/json”}]},{“id”:”986″,”knownName”:{“en”:”Michael Houghton”},”fullName”:{“en”:”Michael Houghton”},”portion”:”1/3″,”sortOrder”:”2″,”motivation”:{“en”:”for the discovery of Hepatitis C virus”,”se”:”för upptäckten av hepatit C-virus”},”links”:[{“rel”:”laureate”,”href”:”https://api.nobelprize.org/2/laureate/986″,”action”:”GET”,”types”:”application/json”}]},{“id”:”987″,”knownName”:{“en”:”Charles M. Rice”},”fullName”:{“en”:”Charles M. Rice”},”portion”:”1/3″,”sortOrder”:”3″,”motivation”:{“en”:”for the discovery of Hepatitis C virus”,”se”:”för upptäckten av hepatit C-virus”},”links”:[{“rel”:”laureate”,”href”:”https://api.nobelprize.org/2/laureate/987″,”action”:”GET”,”types”:”application/json”}]}] |
| 2021 | 2021-10-04 | 10000000 | The Nobel Prize in Physiology or Medicine | [{“id”:”997″,”knownName”:{“en”:”David Julius”},”fullName”:{“en”:”David Julius”},”portion”:”1/2″,”sortOrder”:”1″,”motivation”:{“en”:”for their discoveries of receptors for temperature and touch”},”links”:[{“rel”:”laureate”,”href”:”https://api.nobelprize.org/2/laureate/997″,”action”:”GET”,”types”:”application/json”}]},{“id”:”998″,”knownName”:{“en”:”Ardem Patapoutian”},”fullName”:{“en”:”Ardem Patapoutian”},”portion”:”1/2″,”sortOrder”:”2″,”motivation”:{“en”:”for their discoveries of receptors for temperature and touch”},”links”:[{“rel”:”laureate”,”href”:”https://api.nobelprize.org/2/laureate/998″,”action”:”GET”,”types”:”application/json”}]}] |
JSON_TABLE transformed each award within the array into a row. Fields within the array are mapped to columns – like award_year and category.
This output is much better – but understanding the Nobel Laureates is still a bit of a challenge. As you can see, there are several laureates for each award – represented by a JSON array. The query below will make a minor modification to the previouis JSON_TABLE query. It will use the nested path clause to expand the nested array – adding rows for each Nobel Laureate:
with prizes as (
-- make the REST call
select nobel_prizes(2015, 2021, 'med') as data
from dual
)
select award_year,
date_awarded,
prize_amount,
category,
laureate_name,
motivation
from prizes p,
json_table(p.data, '$.nobelPrizes[*]'
columns (
award_year varchar2(4) path '$.awardYear',
date_awarded varchar2(12) path '$.dateAwarded',
prize_amount number path '$.prizeAmount',
category varchar2(100) path '$.categoryFullName.en',
laureates clob format json path '$.laureates',
nested path '$.laureates[*]'
columns (
laureate_name varchar2(100) path '$.knownName.en',
motivation varchar2(100) path '$.motivation.en'
)
)
) jt
;
| AWARD_YEAR | DATE_AWARDED | PRIZE_AMOUNT | CATEGORY | LAUREATE_NAME | MOTIVATION |
|---|---|---|---|---|---|
| 2019 | 2019-10-07 | 9000000 | The Nobel Prize in Physiology or Medicine | William G. Kaelin Jr | for their discoveries of how cells sense and adapt to oxygen availability |
| 2019 | 2019-10-07 | 9000000 | The Nobel Prize in Physiology or Medicine | Sir Peter J. Ratcliffe | for their discoveries of how cells sense and adapt to oxygen availability |
| 2019 | 2019-10-07 | 9000000 | The Nobel Prize in Physiology or Medicine | Gregg L. Semenza | for their discoveries of how cells sense and adapt to oxygen availability |
| 2020 | 2020-10-05 | 10000000 | The Nobel Prize in Physiology or Medicine | Harvey J. Alter | for the discovery of Hepatitis C virus |
| 2020 | 2020-10-05 | 10000000 | The Nobel Prize in Physiology or Medicine | Michael Houghton | for the discovery of Hepatitis C virus |
| 2020 | 2020-10-05 | 10000000 | The Nobel Prize in Physiology or Medicine | Charles M. Rice | for the discovery of Hepatitis C virus |
| 2021 | 2021-10-04 | 10000000 | The Nobel Prize in Physiology or Medicine | David Julius | for their discoveries of receptors for temperature and touch |
| 2021 | 2021-10-04 | 10000000 | The Nobel Prize in Physiology or Medicine | Ardem Patapoutian | for their discoveries of receptors for temperature and touch |
This is what we want. We can now easily see each of the awards and its laureates. To save this result into a database table, simply add a CREATE TABLE as SELECT to the beginning of the query:
create table nobel_prize_winners as
with prizes as (
-- make the REST call
select nobel_prizes(2015, 2021, 'med') as data
from dual
)
select award_year,
date_awarded,
prize_amount,
category,
laureate_name,
motivation
from prizes p,
json_table(p.data, '$.nobelPrizes[*]'
columns (
award_year varchar2(4) path '$.awardYear',
date_awarded varchar2(12) path '$.dateAwarded',
prize_amount number path '$.prizeAmount',
category varchar2(100) path '$.categoryFullName.en',
laureates clob format json path '$.laureates',
nested path '$.laureates[*]'
columns (
laureate_name varchar2(100) path '$.knownName.en',
motivation varchar2(100) path '$.motivation.en'
)
)
) jt
;
There are more sophisticated approaches to tackling this example which would apply to larger data sets returned by the REST endpoint. SQL table functions allow you to add the function to the FROM clause of the query. Within that function, you’ll want to implement a pipeline to stream pages back to the select statement instead of processing the entire result at one time (a good topic for a blog post!).
Summary
With data coming from so many different types of sources, a powerful SQL engine is a real difference maker. It greatly simplifies application deployments – allowing you to solve problems directly from SQL and/or PLSQL functions without needing to deploy other data processing tiers. There are no connectivity challenges, debugging is less complex and it allows for a more efficient, iterative development process.
Try this example on your own Autonomous Database instance. It’s easy to swap in other data sources for the Nobel Prize example. If you don’t have ADB, then sign up for free. Everything we’ve done here can be done on the Always Free tier.
