In the rapidly evolving field of generative AI, Mistral AI recently unveiled the Mixtral 8X7B. This model, a Sparse Mixture of Experts (SMoE) architecture, shares similarities with the Mistral 7B but introduces a novel feature: it combines eight “expert” models into one. For a deeper understanding of the sparse mixture of experts, consider exploring this blog from Huggingface. The Mixtral 8X7B outshines the Llama 2-70B in most benchmarks, offering inference speeds that are six times faster. Often likened to a more compact version of GPT-4, the Mixtral 8X7B employs a Mixture of Experts (MoE) framework with eight experts, each comprising 111B parameters, along with 55B shared attention parameters, totaling 166B parameters per model. This strategic design enables only two experts to participate in the inference for each token, marking a trend towards more streamlined and targeted AI processing. A standout feature of Mixtral is its capacity to handle an extensive 32,000-token context, adeptly managing intricate tasks. Its multilingual support extends to English, French, Italian, German, and Spanish, making it a valuable tool for a diverse global developer community. The pre-training phase of Mixtral involves data from the open web and employs a concurrent training method for both experts and routers, ensuring the model is not only expansive in its parameter scope but also finely attuned to the subtleties of its extensive data exposure. Next, we will delve into the hardware requirements for this model.
Sizes
| Name |
Number of parameters |
Number of active parameters |
Min. GPU RAM for inference (GB) |
| Mistral-7B-v0.2 |
7.3B |
7.3B |
16 |
| Mistral-8X7B-v0.1 |
46.7B |
12.9B |
100 |
source: https://docs.mistral.ai/models/
For the step-by-step walkthrough of the process, see the AI samples GitHub repository
About the Dataset
The dataset is taken from Pubmed. The PubMed QA dataset is a specialised resource designed for the development and evaluation of automatic question-answering systems in the biomedical domain. This dataset is particularly valuable because it is derived from PubMed, a free search engine accessing primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics. The PubMed QA dataset consists of a collection of question and answer pairs, which are meticulously curated to reflect a wide range of medical and healthcare topics. These pairs are often accompanied by relevant context or reference information extracted from scientific publications. The dataset is instrumental for training and testing natural language processing algorithms, especially those focused on understanding and responding to complex, domain-specific inquiries. Its use is critical in advancing the capabilities of AI in healthcare, enabling the development of sophisticated tools that can assist in medical research, diagnostics, and patient care by providing accurate and contextually relevant answers to medical queries. We would be making use of the python.
High Level Solution Overview
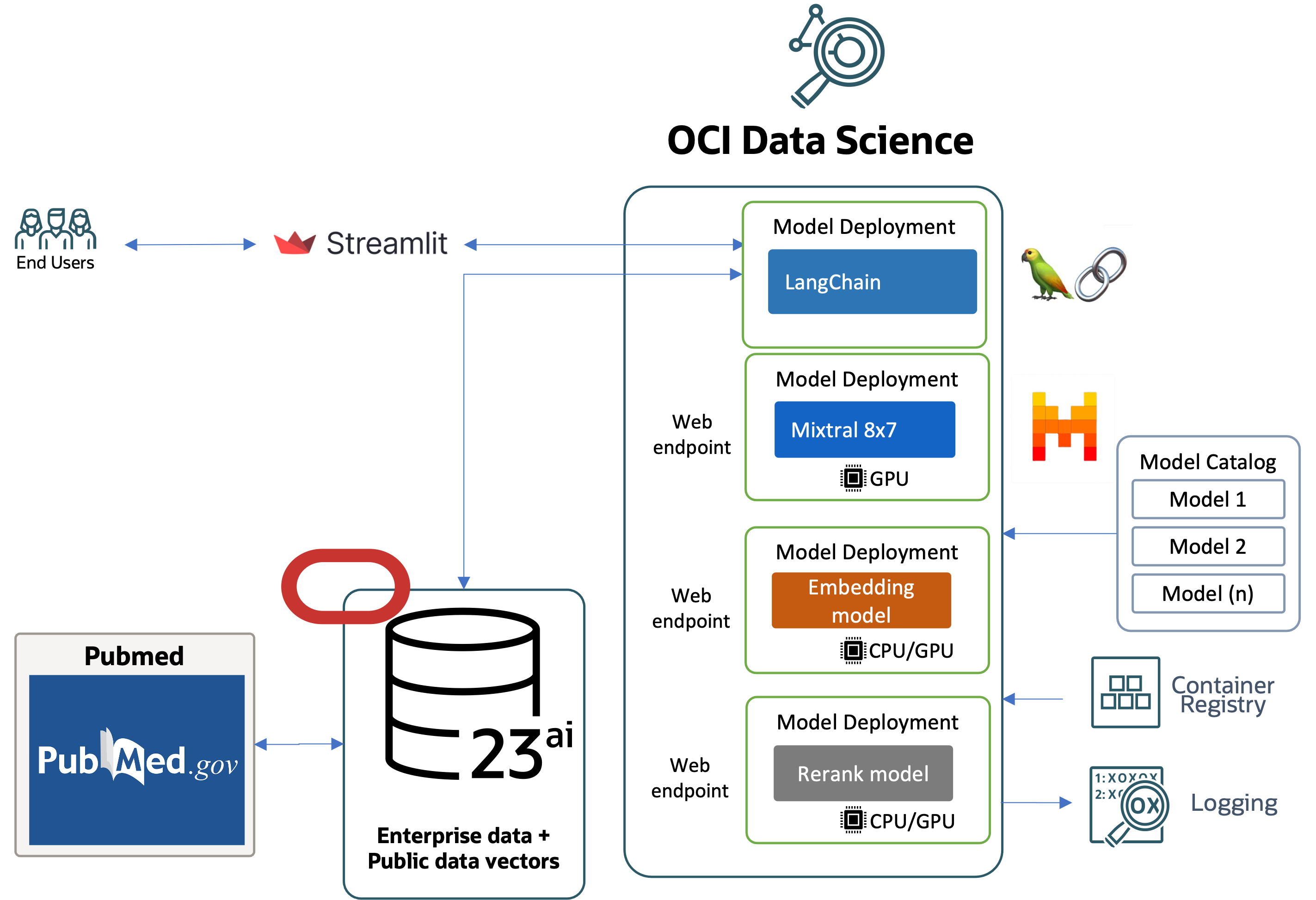
Image: High level overview of a RAG Pipeline
Steps
In this scenario the steps that would be involved includes step 0 which is fetching documents from pubmed and vectorize the data into Vector db such as Oracle 23AI. The diagram illustrates the process flow for a healthcare chatbot system that integrates various technologies and data sources to generate responses:
- Streamlit Interface: Streamlit captures user inputs and sends them to the backend model deployments. The system vectorizes the user query using the embedding model deployment via the web endpoint
- Query processing: The vectorized query is sent to the Oracle DB 23ai, which contains enterprise data, and data fetched from the pubmed datasets which are already stored as embeddings.
- Data Retrieval – The system retrieves relevant data from PubMed based on the vectorized query
- Response Processing – The retrieved data is processed by the Oracle DB 23ai to generate a response.
- Ranking – The response is ranked using the re-rank model deployment via the web endpoint.
- LLM Query – The ranked response is sent to the LangChain model deployment via the web endpoint for language model processing.
- LLM Generated Response – The LangChain model generates a refined response based on the ranked data.
- Logging and Monitoring: The system employs logging mechanisms to track the performance and outputs of the model deployments for monitoring and troubleshooting purposes.
- Feedback to User: Processed outputs from the model deployments are returned to the Streamlit interface, where they are displayed to the user, completing the interaction loop
This process integrates public data sources like PubMed and employs model management and deployment strategies, ensuring that responses are accurate, relevant, and up-to-date.
Pre-requisites
The key prerequisites that you would need to setup before you can proceed to run the distributed fine-tuning process on Oracle Cloud Infrastructure Data Science Service.
- Policies – the following policies are needed
- Configure custom subnet – with security list to allow ingress into any port from the IPs originating within the CIDR block of the subnet. This is to ensure that the hosts on the subnet can connect to each other during distributed training.
- Create an object storage bucket – to save the documents which are provided at time of ingestion in vector DB.
- Set the policies – to allow the OCI Data Science Service resources to access object storage buckets, networking and others
- Access token from HuggingFace to download Llama2 model. To fine-tune the model, you will first need to access the pre-trained model. The pre-trained model can be obtained from Meta or HuggingFace. In this example, we will use the HuggingFace access token to download the pre-trained model from HuggingFace (by setting the HUGGING_FACE_HUB_TOKEN environment variable).
- Log group and log from logging service. This will be used to monitor the progress of the training
- Go to the OCI Logging Service and select Log Groups
- Either select one of the existing Log Groups or create a new one
- In the log group create two Log, one predict log and one access log, like:
- Click on the Create custom log
- Specify a name (predict|access) and select the log group you want to use
- Under Create agent configuration select Add configuration later
- Then click Create agent configuration
- Notebook session – used to initiate the distributed training and to access the fine-tuned model
Install the latest version of oracle-ads - pip install oracle-ads[opctl] -U
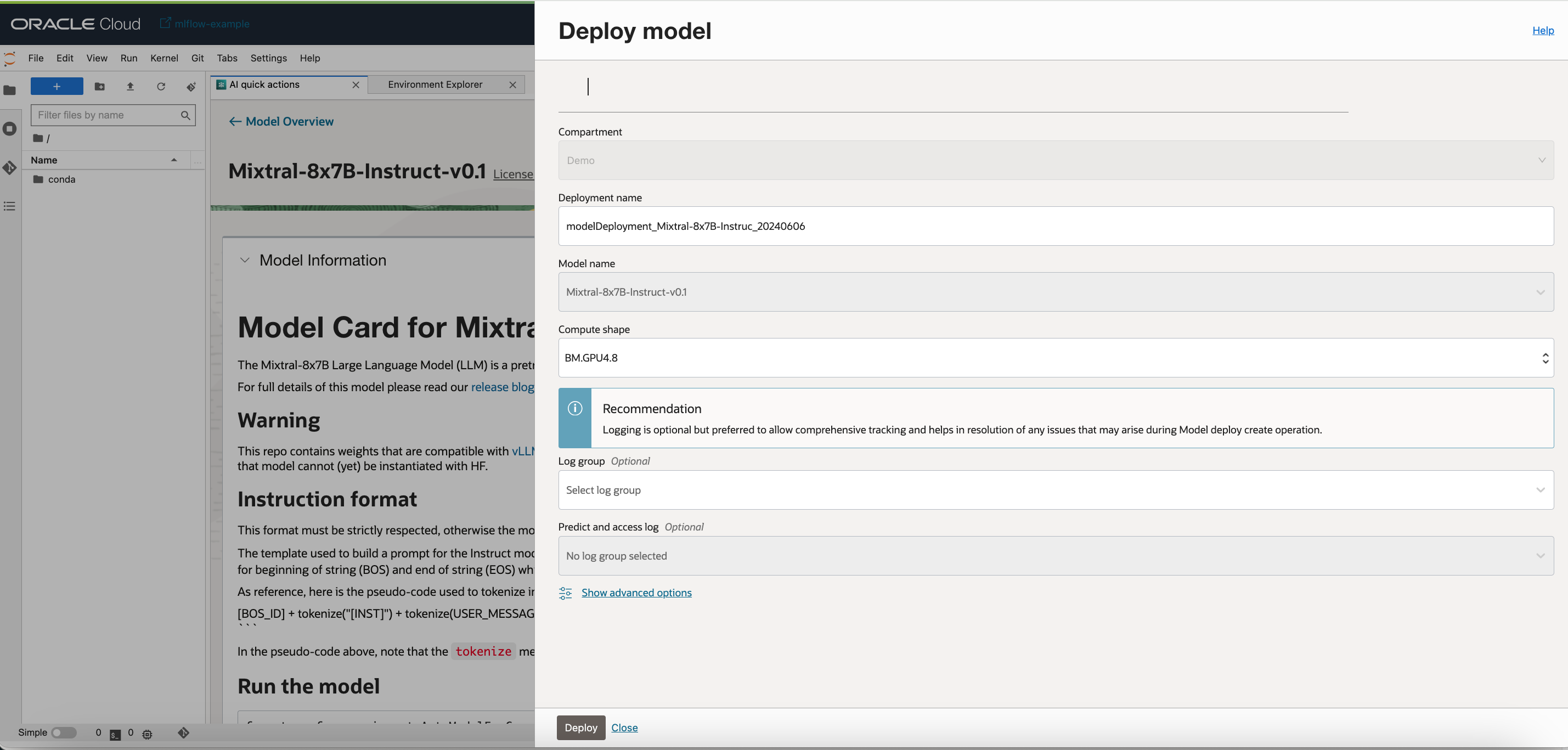
Step 1 : Deploying the Mixtral 8X7B Model
We would be leveraging AI quick actions Model deployment feature, to deploy Mixtral 8X7B with a few clicks. It is essential to have more than 100GB of GPU memory and a container size of more than 120GB in size. The AI Quick actions model deployment would aid users to deploy the Mixtral 8X7B with a few clicks and provide users with an endpoint.
Note: To estimate model memory needs, Hugging Face offers a Model Memory Calculator. Furthermore, for insights into the fundamental calculations of memory requirements for transformers, Eleuther has published an informative article on the subject.
Step 2: Setting up the Oracle 23AI database
We will be utilizing the latest features of Oracle 23AI, specifically the AI Vector Search, for our use case. In this scenario, we will import the embeddings generated from the PubMed dataset into Oracle 23AI. When a user queries using the RAG, the response will be enhanced by supplying the LLMs with additional context. the below diagram shows some of benefits of using Oracle 23AI:
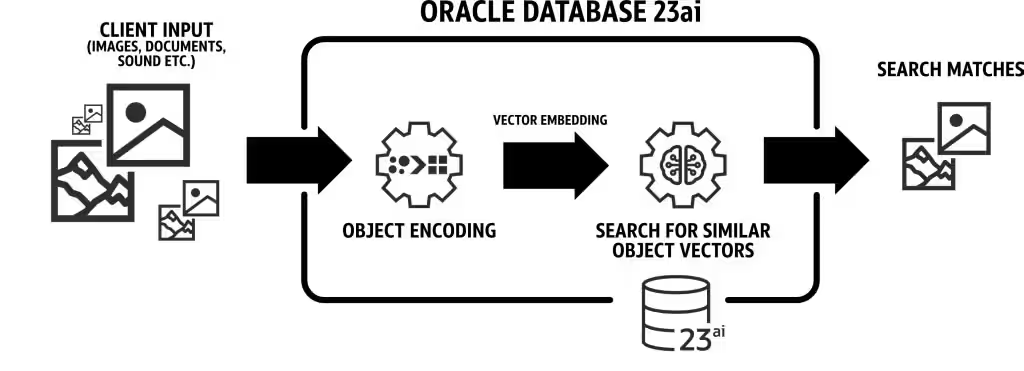
Figure: A illustration of Oracle 23AI Search Feature (source)
This will augment their knowledge, leading to responses that are more accurate and pertinent to the customer’s inquiries To setup Oracle 23AI you can following the options mentioned below:
- Using a container instance as mentioned here
- using a python client
We will be using a hosted instance of Oracle 23AI to demonstrate the powerful features of Vector search.
Step 3: Initialize Oracle 23AI with Langchain
Oracle 23AI has integrates smoothly with LangChain, and you can actually use Oracle23AI within LangChain via the VectorDBQA class. The initial step is to compile all the documents that will act as the foundational knowledge for our LLM. Imagine we place these in a list titled docs. Each item in this docs list is a string containing segments of paragraphs.
Oracle 23AI Initialisation
Below code snippet helps set connection to your Oracle 23AI vector database.
Oracle 23 AI Connection
import oracledb
import sys
password = "<USER_SET_PWD>"
conn=oracledb.connect(
config_dir="DIRECTORY_OF_WALLET",
user="<USER_NAME>",
password=password,
dsn="DSN_NAME",
wallet_location="DIRECTORY_OF_WALLET",
wallet_password=password)
Getting Documents and Text Chunking
Download PubMed Dataset and chunk them before uploading to Oracle 23AI.
Get Documents and Text chunking
# Utilise Langchain to download
from langchain_community.document_loaders import PubMedLoader
loader = PubMedLoader("diabetes", load_max_docs=2000) # Choose Topics
docs = loader.load()
# Text Chunking
from langchain_community.document_loaders.oracleai import OracleTextSplitter
from langchain_core.documents import Document
# split by default parameters
splitter_params = {"normalize": "all"}
""" get the splitter instance """
splitter = OracleTextSplitter(conn=conn, params=splitter_params)
count = 0
list_chunks = []
for doc in docs:
print(count)
count += 1
chunks = splitter.split_text(doc.page_content)
list_chunks.extend(chunks)
""" verify """
print(f"Number of Chunks: {len(list_chunks)}")
Load Embeddings and Upload to Vector Store
We can load the embeddings locally or on Oracle 23 AI. Steps to both are shared below, we use local embeddings by default:
from langchain_community.embeddings import HuggingFaceEmbeddings
# Locally stored Embeddings
embeddings = HuggingFaceEmbeddings(model_name="MODEL NAME")
########### OR ############
from langchain_community.embeddings.oracleai import OracleEmbeddings
# please update with your related information
# make sure that you have onnx file in the system
onnx_dir = "DIRECTORY TO ONNX FILE"
onnx_file = "ONNX FILE NAME"
model_name = "MODEL NAME"
try:
OracleEmbeddings.load_onnx_model(conn, onnx_dir, onnx_file, model_name)
print("ONNX model loaded.")
except Exception as e:
print("ONNX model loading failed!")
sys.exit(1)
# Connect to vector DB
vector_store_dot = OracleVS(
client=conn,
embedding_function=embeddings,
table_name="Documents_DOT",
distance_strategy=DistanceStrategy.DOT_PRODUCT,
)
# Upload to texts
vector_store_dot.add_texts(list_chunks)
# Retrieve Answers
answers = vector_store_dot.similarity_search("diabetes in africa", 20)
Step 4: Connect to Mixtral8x7B AQUA LLM
# set authentication mechanism
import ads
ads.set_auth(auth="api_key")
oci_md = OCIModelDeploymentVLLM(
endpoint="ENDPOINT",
model="/opt/ds/model/deployed_model"
)
Step 5 : Setting up RAG
The following RAG architecture, would consist of the following steps:
template = """You are an assistant to the user, you are given some context below, please answer the query of the user with as detail as possible
Context:\"""
{context}
\"""
Question:\"
{question}
\"""
Answer:"""
qa_prompt = PromptTemplate.from_template(template)
# User Query
question = "Tell me something about Diabetest in africa"
docs = vector_store_dot.similarity_search(question, 5)
prompt = template.format(context=docs, question=question)
llm_answer = oci_md.generate(prompts=[prompt])
We use the prompt template and QA chain provided by Langchain to make the chatbot, this helps in passing the context and question directly to the LLM.
Step 6 : Building an Streamlit chatbot app to host RAG
Setting up Compute Instance
- Create a compute instance with public subnet with internet gateway.
-
Create a dynamic group and add the compute instance ocid to it.
-
Provide the following in policies for the dynamic group
allow group data-science-model-deployments to manage data_science_projects in compartment <datascience_hol>
Hosting Streamlit application
Authentication to access the OCI Model deployment service, include this in the streamlit application while sending request to Model Deployment.
from oci.auth import signers
config = {"region": <YOUR_REGION>}
signer = signers.InstancePrincipalsSecurityTokenSigner()
endpoint = <MD_ENDPOINT>
prompt = <USER_PROMPT>
response = requests.post(endpoint, data=prompt, auth=signer, headers=headers, timeout=200)
For requirements and the Streamlit Python code, see the the GitHub repository.
Benchmarking Results
Let’s compare the outputs from directly asking the LLM vs using RAG with LLM. The LLM originally gives the following response:
Diabetes is a serious health problem in Africa, where it affects millions of people. According to the World Health Organization (WHO), Africa has the highest burden of diabetes in the world, with an estimated 18.3 million people living with the disease in 2019. This represents about 60% of the global total of people living with diabetes.There are several factors that contribute to the high prevalence of diabetes in Africa, including:
1. Urbanization and lifestyle changes: As people move from rural areas to cities, they often adopt unhealthy lifestyles, such as a diet high in sugar and fat, and a lack of physical activity. This can increase the risk of developing diabetes.
2. Obesity: Many Africans are becoming increasingly overweight and obese, which is a major risk factor for developing diabetes.
3. Lack of access to healthcare: In many parts of Africa, people do not have access to basic healthcare services, including diabetes diagnosis and treatment.
With the RAG pipeline, the LLM produces the following response:
Africa has a high prevalence of diabetes, with a pooled prevalence of 8.3% among adults in sub-Saharan Africa, which is higher than the global average of 7.8%. The prevalence of diabetes varies widely across the region, with the highest rates found in north Africa and the Middle East (10.3%), followed by sub-Saharan Africa (8.3%), and the lowest rates found in south Africa (6.8%).
The study found that non-autoimmune diabetes is more common in underweight individuals in sub-Saharan Africa, characterized by the absence of visceral adiposity, reduced late-phase insulin secretion, and greater insulin deficiency. The study also found that the prevalence of poor sleep quality is high among diabetes patients in Ethiopia, with almost half of the patients having poor sleep quality.
The burden of diabetes in Africa is expected to increase in the future, with the largest shift towards morbidity and away from premature death forecasted for sub-Saharan Africa.
Comparison: The answer by RAG is much more detailed as compared to the direct answer, as seen through multiple data points and even talking about specific regions having issues in Africa.
Conclusion
The development of an advanced AI healthcare chatbot using Mixtral 8X7B, Oracle 23AI RAG, LangChain, and Streamlit represents a significant stride in the integration of cutting-edge AI technologies within the healthcare field. This guide has walked through the entire process, from understanding the architecture of models like Mixtral 8X7B to deploying on OCI AI Quick Actions. By utilizing PubMed datasets and implementing effective hardware setups, we have demonstrated how to enhance the capabilities of AI in providing accurate, contextually relevant answers to complex medical queries. The step-by-step approach ensures that developers can replicate and customize this solution, paving the way for more sophisticated and reliable AI-driven healthcare solutions. As AI continues to evolve, such integrations can be crucial in driving innovation and improving patient care globally
Try Oracle Cloud Free Trial! A 30-day trial with US$300 in free credits gives you access to Oracle Cloud Infrastructure Data Science service. For more information, see the following resources:
- Full sample, including all files in OCI Data Science sample repository on GitHub.
- Visit our service documentation.
- Watch our tutorials on our YouTube playlist.
- Try one of our LiveLabs. Search for “data science.”
- Got questions? Reach out to us at ask-oci-data-science_grp@oracle.com
If you want to suggest specific models to add to AI Quick Actions email the OCI Data Science group. For more information on how to use AI Quick Actions, go to the Oracle Cloud Infrastructure Data Science YouTube playlist to see a demo video of AI Quick Actions, and find our technical documentation, and see our Github repository with tips and examples.
Acknowledgements
- Announcing AI Quick Actions : https://blogs.oracle.com/ai-and-datascience/post/ai-quick-actions-in-oci-data-science
- Announcing Oracle Database 23ai : General Availability : https://blogs.oracle.com/database/post/oracle-23ai-now-generally-available




