Many customers are keep asking me about “default” (single) compression codec for Hadoop. Actually answer on this question is not so easy and let me explain why.
Bzip2 or not Bzip2?
In my previous blogpost I published results of the compression rate for some particular compression codecs into Hadoop. Based on those results you may think that it’s a good idea to compress everything with bzip2. But be careful with this. Within the same research, I noted that bzip2 actually has on average 3 times worse performance than Gzip for querying (decompress) and archive (compress) data (it’s not surprising based on the complexity of algorithm). Are you ready to sacrifice performance? I think it will depend on the compression benefits derived from bzip2 and the frequency of querying this data (compression speed is not so import after data is stored in Hadoop systems since you usually compress data once and read it many times). On average, bzip2 is 1.6 times better than gzip. But, again my research showed that sometimes you can achieve 2.3 times better compression, while other times you may gain only 9% of the disk space usage (and performance is still much worse compared to gzip and other codecs). Second factor to keep in mind is the frequency of data querying and your performance SLAs. If you don’t care about query performance (don’t have any SLAs) and you select this data very rarely – bzip2 could be good a candidate. Otherwise consider other options. I encourage you to benchmark your own data and decide for yourself “Bzip2 or not Bzip2”.
Gzip or not gzip?
Ok, let’s imagine that for some reasons you have decided against bzip2 codec (for performance reasons or it just doesn’t bring any compression benefits). Gzip is now going to be your next candidate as the default codec. But wait! It’s not splittable. What does this actually mean? Let me briefly explain. As an example, let’s assume we have 1GB file on the cluster with 256MB blocks. This means that the file is split into 4 blocks. Let’s imagine that we have 2 servers and 2 CPU cores (+ some memory, but let’s ignore this) on each server for processing.
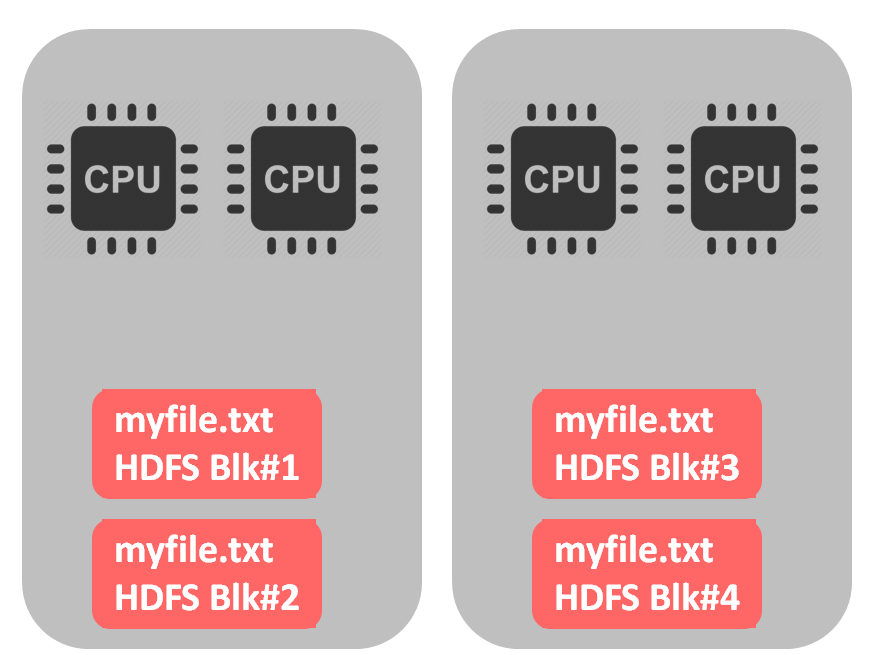
If the file is splittable, we are able to use all cpus for parallel processing, as shown on picture below:
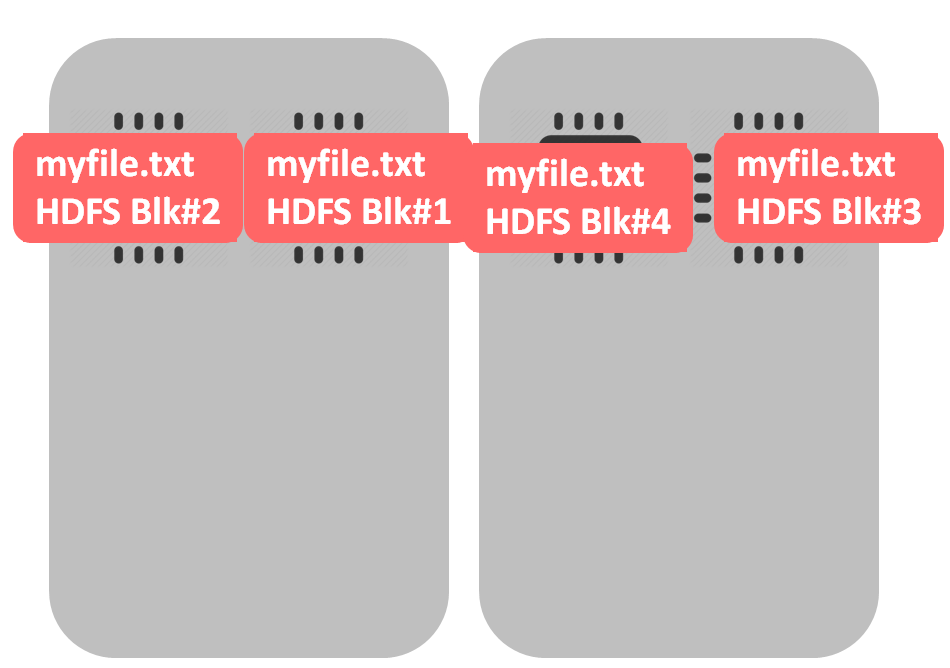
What would be the case of a non-splittable file – like gzip? The file would be split into 4 HDFS blocks (during data loading into HDFS), like in example above. The difference begins when we start processing this file – all blocks would be processed by a single cpu. In our example one core will be occupied while 3 others are idle.
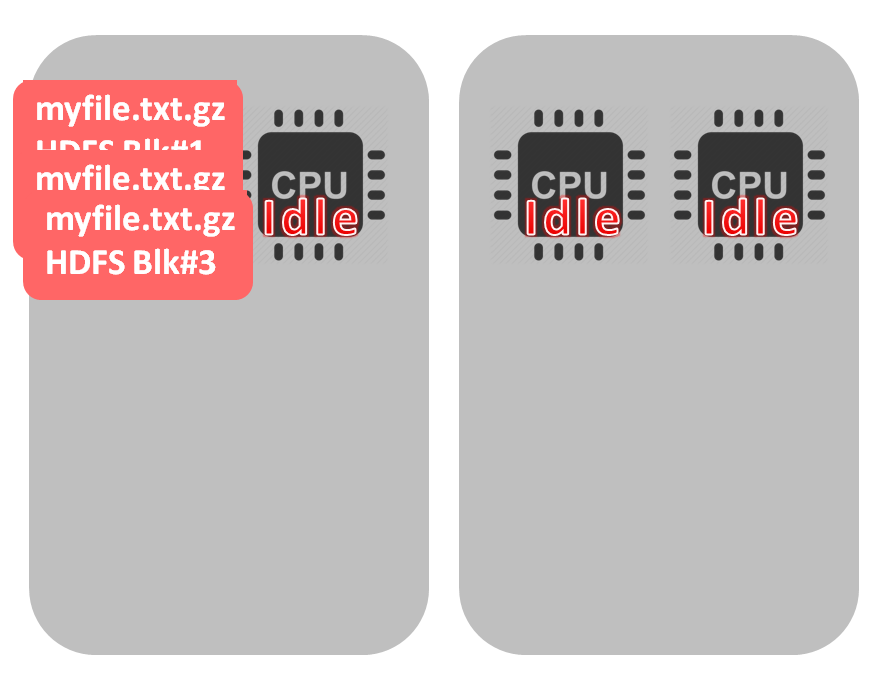
Not good, right? But non-splittable files – like gzip – are always a problem. For example, if the file size approximates the HDFS block size (or even less) it’s fine (because we don’t need to distribute the file precessing across multiple cores). One file, one block, one cpu. If you have data source that provides gzip files – then you probably are unable to influence the files size. But, oftentimes files are produced by MapReduce (like hive, for example) or Spark jobs and the number of Reducers or Mappers determines the number of files and their size. For example, experimenting with mapreduce.job.reduces (defines the number of reducers and as a consequence the number of the output files) parameter I ran the same MapReduce job and analyzed the number of files and the average size:
2 Reducers – 2 Files – 5.9GB each (it’s bad, because one CPU will handle 5.9 GB file)
994 Reducers – 497 Files – 12.5MB (it’s also bad, because we will have so many small files in HDFS)
497 Reducers (default) – 497 Files – 24.7MB (better than previous example, but file size is still small)
48 Reducers – 48 Files – 255MB (it’s good. File size seems close to the block size)
This case shows that by setting proper number of output jobs (files) you could handle proper size of the gzip file.
LZ4, Snappy, LZO and others
On Big Data Appliance, Gzip performance is usually comparable with Snappy or LZ4 or maybe a bit worse. But, unlike Gzip, LZ4 and Snappy are splittable. If you are not able to control the number of reducers or you just don’t want to do so (there are processing performance implications), consider using Snappy or LZ4. They are splittable and have a bit better performance. The one thing that you must check is compatibility with the technology/applications that you are using. For example, Impala doesn’t support LZ4, which is usually a bit better from compression ratio and performance perspective than Snappy. However, Impala does support Snappy. Not all applications support all file formats (like sequencefiles, RC, ORC, parquet) and all compression codecs (like bzip2, gzip, lz4, lzo, snappy). I have seen many times when the combination of RC/gzip had best compress/performance rate, but because of incompatibility with some specific applications, customers used textfile/snappy combination instead.
Any other options?
This research investigated 5 major compression codecs available in many hadoop distributions: bzip2, gzip, lz4, lzo, snappy. But am I limited by these 5 codecs? Generally speaking, the answer is no. You could implement or reuse already implemented algorithms. Like an example, consider the LZMA algorithm. It has a compression rate similar to bzip2 (or even better), but is much faster for decompression (speed of decompression is comparable with gzip). All this sounds perfect – but what is disadvantage? The main disadvantage is compression speed. It’s very very slow. But if you use Hadoop as an extension of the data warehouse, you may only offload and compress data once (and therefore pay the performance penalty only once) and then use advantages of LZMA (compression rate on the bzip2 level, performance decompression comparable with gzip). Let review an example of how to install this compression codec for hive and how to use it:
1) Download
jar file. For example, from here
2) Put
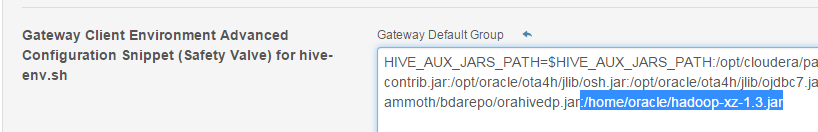
it in some directory on all servers, like /home/oracle/hadoop-xz-1.3.jar
Add this directory in HIVE_AUX_JAR
with Cloudera Manager (clusters -> hive -> configuration):
3) Add
this codec in HDFS parameters (clusters -> hdfs -> configuration):
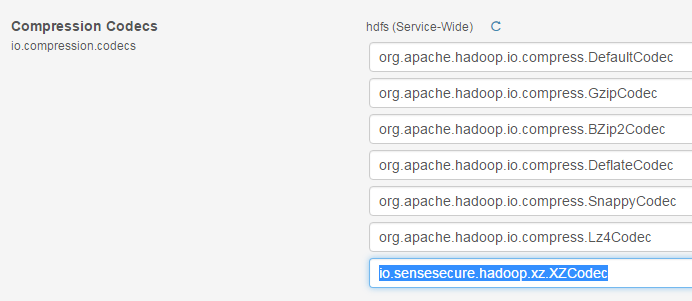
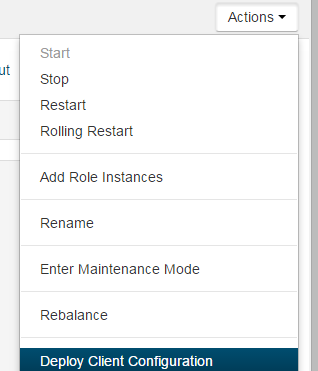
4) Deploy
new config by Cloudera Manager:
5) For
compress data run query like this:
hive> set hive.exec.compress.output=true;
hive> set
io.seqfile.compression.type=BLOCK;
hive> set
mapreduce.output.fileoutputformat.compress.codec=io.sensesecure.hadoop.xz.XZCodec;
hive> CREATE TABLE test_table_lzma ROW
FORMAT DELIMITED FIELDS TERMINATED BY “,” LINES TERMINATED BY
“\n” STORED AS TEXTFILE
LOCATION “/tmp/ test_table_lzma” as select * from some_table;
Enjoy your lzma! I did quick test with
some dataset and got pretty good results:
| bzip2 |
gzip |
lzma |
|
| Time of the compression, Minutes |
36.5m |
14.3m |
97.6m |
| Data size after compression (originally 370GB of uncompressed data), GB |
64 |
50 |
48 |
| Time of decompression (select query), Seconds |
1105 |
462 |
501 |
Unfortunately, right now the lzma codec is not supported by many distributions of Hadoop. As a result, if you experience a problem with this compression codec, support would not be able to help you. So, choose wisely!
