NVIDIA Clara Parabricks Pipelines are an accelerated computational framework supporting genomics applications by providing quick and accurate genome analysis tools, supporting germline, somatic, and RNA workflows. The solution uses an NVIDIA GPU and speeds up analysis of whole genomes from days to under an hour, starting with a fastq file and generating a VCF. This VCF has a 99.99% accuracy and precision when compared to the GATK4 baseline which NVIDIA Parabricks. To demonstrate the increased performance of running Parabricks Pipelines on the Oracle Cloud Infrastructure (OCI) BM.GPU4.8 instance, we ran the germline pipeline using the dataset ERR194147 from the European Nucleotide Archive. This dataset consists of two files, 48 GB and 49 GB, and each contain over 780,000 reads.
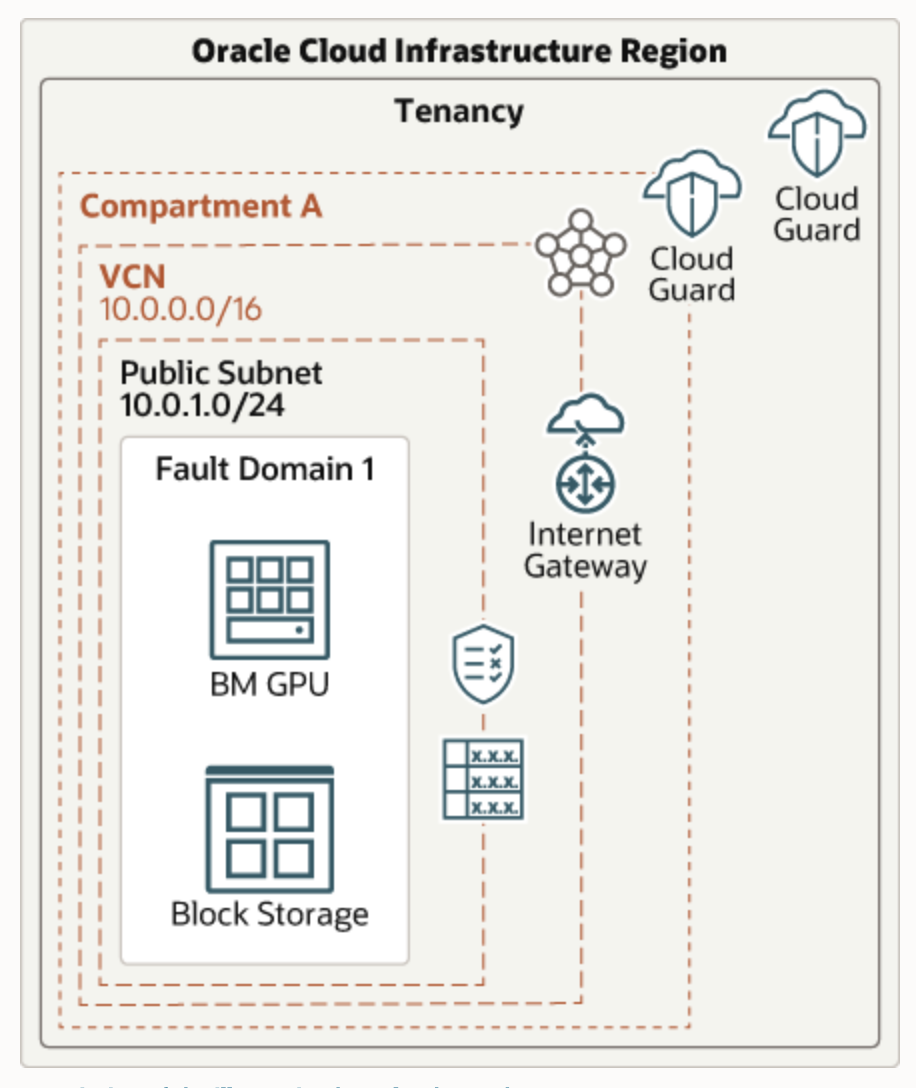
For the environment set up, we had a GPU node with Block Storage deployed in a VCN with a public subnet and internet gateway. The following diagram illustrates this reference architecture. To learn more about this reference architecture, see Deploy genomics applications framework and NVIDIA Clara Parabricks.
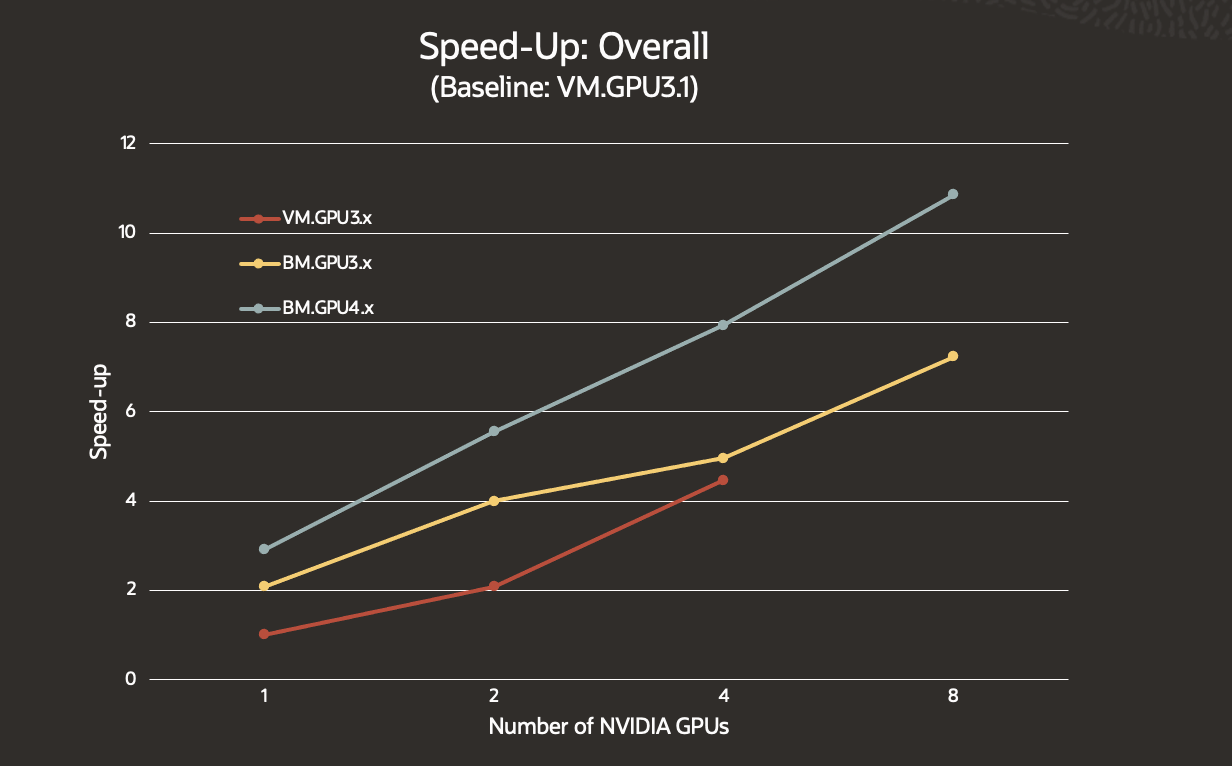
The test compared the NVIDIA V100 Tensor Core GPU-enabled VM.GPU3.1, VM.GPU3.2, VMGPU3.4, and the BM.GPU3.8 instances with the new BM.GPU4.8 instance powered by the NVIDIA A100 Tensor Core GPU. The BM.GPU4.8 had on average 32% greater overall speed-up compared to the BM.GPU3.8.
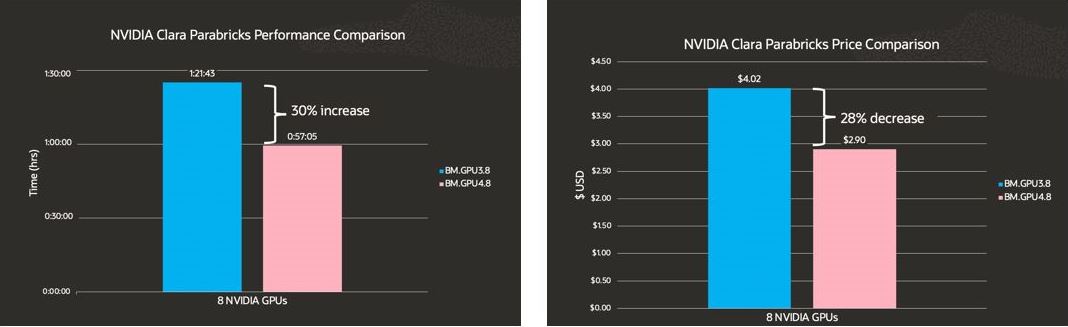
Comparing Parabricks Pipelines on BM.GPU4.8 ($3.05 GPU/hour) against BM.GPU3.8 ($2.95 GPU/hour), you can significantly increase performance while reducing costs. Besides NVIDIA Clara Parabricks, you can get better performance on other molecular dynamic simulation workloads.
Try it yourself on OCI Resource Manager by clicking 
Best Performance for your Molecular Dynamic Simulation Workloads
In addition to NVIDIA Clara Parabricks, you can also get better performance for your Molecular Dynamic (MD) Simulation workloads. MD simulations can be used to analyze the physical movements in atoms or molecules and perform nucleotide and genomic sequencing. Many MD simulations are very computationally demanding and can be accelerated by running on GPUs. Using the latest, most powerful NVIDIA A100 Tensor Core GPUs, performance for applications like Clara Parabricks Pipelines, GROMACS, and NAMD increase substantially. And MD simulations aren’t the only workloads to benefit from the powerful A100 GPUs [See NVIDIA A100 Tensor Core GPU Bare Metal Performance in Oracle Cloud Infrastructure].
Running GROMACS on OCI
GROMACS is a molecular dynamics package primarily designed for biochemical molecules, but because of its ability to quickly calculate non-bonded interactions, many groups also use it for research on non-biological systems. We ran benchmarks on the GROMACS benchmark using the benchPEP data set and saw a 38% increase in performance from BM.GPU3.8.
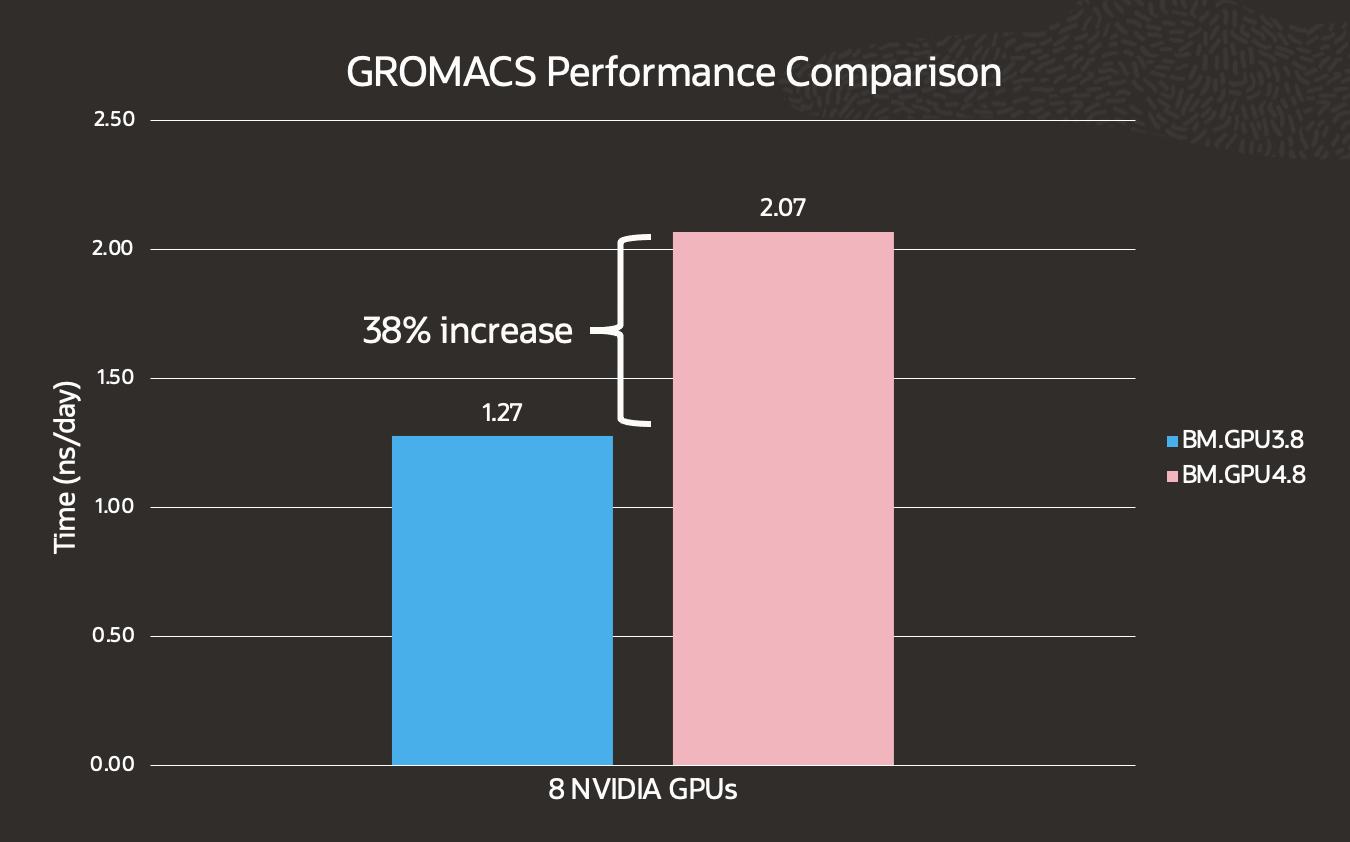
*A higher ns/day indicates better performance
Try it yourself on OCI Resource Manager by clicking 
Running NAMD on OCI
NAMD is a parallel, object-oriented molecular dynamics code designed for large biomolecular systems. We used the standard NAMD benchmark with the ApoA1 model and saw a 32% performance increase.
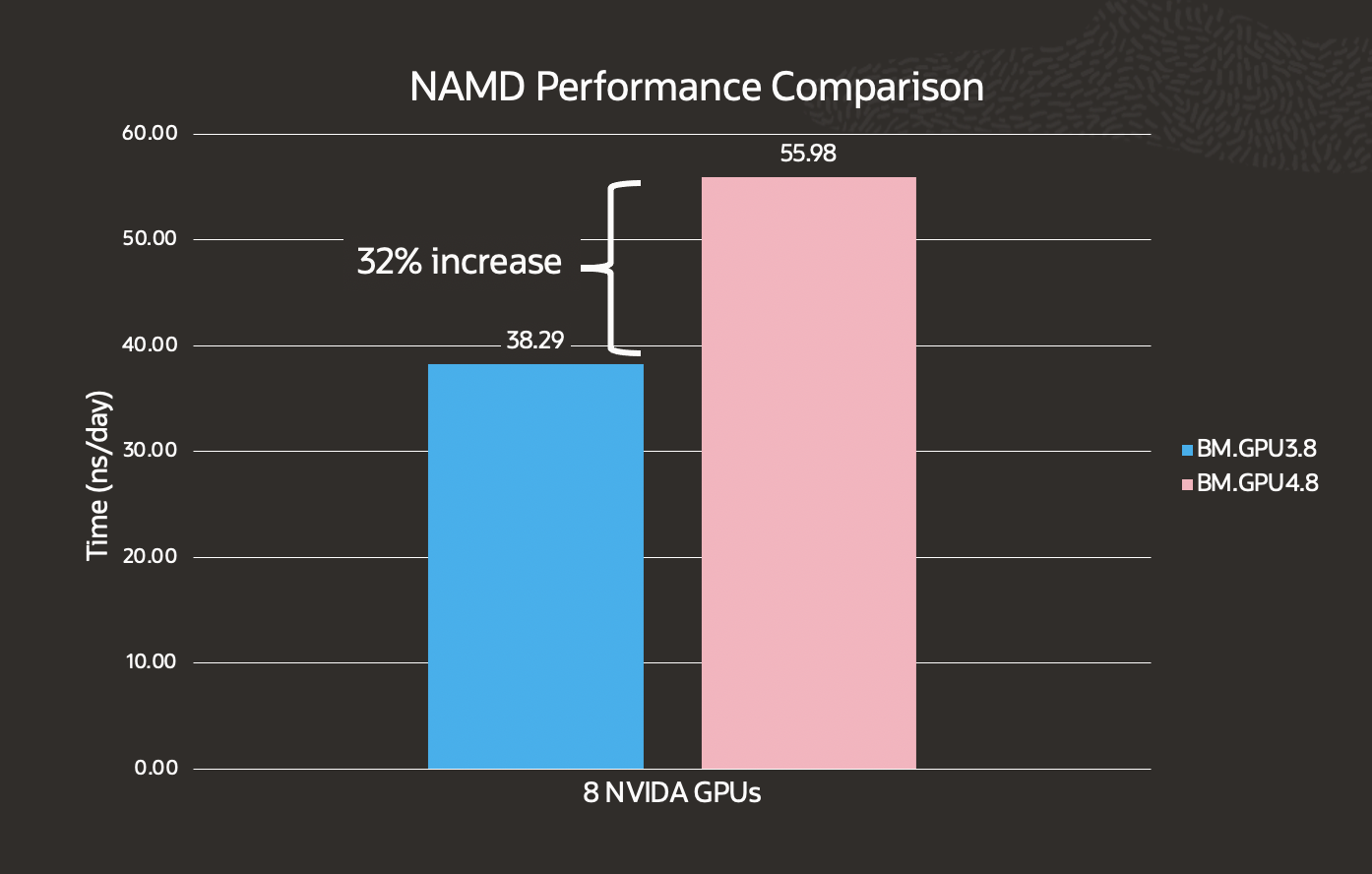
*A higher ns/day indicates better performance
Regardless of which life science simulation application you run, the NVIDIA A100 Tensor Core GPU on OCI can bring you better performance for the best price in the cloud with around 30% performance increase.
Try it yourself on OCI Resource Manager by clicking 
Get Started Today
Start your 30-day free trial and get access to a wide range of Oracle Cloud Infrastructure services for 30 days, including BM.GPU4.8 and BM.GPU3.8 shapes.
Are you a researcher and looking for extra credits? Use OCI for your research by signing up for a Research Cloud Starter Award from Oracle for Research, $1,000 credit towards a variety of cloud storage, database, and service offerings.
To get more details on how to run these applications please refer to the following GitHub repositories: Parabricks, GROMACS, and NAMD. In addition, if want to learn more about running molecular dynamic simulations, check out our life sciences page.
